P3k14C: A synthetic global database of archaeological radiocarbon dates
Source:vignettes/articles/p3k14c.Rmd
p3k14c.Rmd
library(p3k14c)
#> Loading required package: magrittr
library(ggplot2)
# options(scipen = 999)
dir.create(here::here("vignettes/articles/figures"),
showWarnings = FALSE,
recursive = TRUE)
dir.create(here::here("vignettes/articles/figures"),
showWarnings = FALSE,
recursive = TRUE)
global_admin1 <-
# Global Admin 1 boundary dataset
"https://www.geoboundaries.org/data/geoBoundariesCGAZ-3_0_0/ADM1/simplifyRatio_25/geoBoundariesCGAZ_ADM1.topojson" %>%
sf::read_sf(crs = 4326)
# Radiocarbon date data
p3k14c_data <-
p3k14c::p3k14c_data %>%
dplyr::count(SiteID,
SiteName,
Continent,
Long,
Lat,
name = "Dates")
# Count dates/sites
p3k14c_data %>%
dplyr::group_by(Continent) %>%
dplyr::summarise(Dates = sum(Dates, na.rm = TRUE),
Sites = dplyr::n()) %>%
na.omit() %>%
dplyr::arrange(dplyr::desc(Dates))
#> # A tibble: 7 × 3
#> Continent Dates Sites
#> <chr> <int> <int>
#> 1 Europe 77144 21184
#> 2 North America 64905 16097
#> 3 Asia 13995 2676
#> 4 Africa 11101 3497
#> 5 South America 7670 2064
#> 6 Australia 3657 1530
#> 7 Central America 1217 99
# Drop non-spatial data for risk analysis
p3k14c_data %<>%
tidyr::drop_na(Long, Lat) %>%
sf::st_as_sf(
coords = c("Long", "Lat"),
crs = "EPSG:4326",
remove = FALSE
) %>%
# Transform to equal earth projection (EPSG 8857)
sf::st_transform("EPSG:8857")
here::here("../p3k14c-data/p3k14c_raw.csv") %>%
readr::read_csv(col_types =
readr::cols(
Age = readr::col_double(),
Error = readr::col_double(),
.default = readr::col_character()
)) %>%
dplyr::group_by(Continent) %>%
dplyr::summarize(
`N Raw Dates (including duplicates)` = dplyr::n()
) %>%
dplyr::left_join(
p3k14c::p3k14c_data %>%
dplyr::group_by(Continent) %>%
dplyr::summarize(
`N Scrubbed Dates` = dplyr::n(),
`Mean Error (years)` = mean(Error, na.rm = TRUE),
`Median Error (years)` = median(Error, na.rm = TRUE)
)
) %>%
dplyr::mutate(`% Dates Retained` =
100 * `N Scrubbed Dates` /
`N Raw Dates (including duplicates)`) %>%
dplyr::select(Continent,
`N Raw Dates (including duplicates)`,
`N Scrubbed Dates`,
`% Dates Retained`,
`Mean Error (years)`,
`Median Error (years)`) %T>%
readr::write_csv(
here::here("vignettes/articles/tables/p3k14c_continental_summary.csv")
)
#> Warning: 6 parsing failures.
#> row col expected actual file
#> 112741 Age a double Bomb C14 '/Users/bocinsky/git/publications/p3k14c/../p3k14c-data/p3k14c_raw.csv'
#> 112750 Age a double Bomb C14 '/Users/bocinsky/git/publications/p3k14c/../p3k14c-data/p3k14c_raw.csv'
#> 112778 Age a double Bomb C14 '/Users/bocinsky/git/publications/p3k14c/../p3k14c-data/p3k14c_raw.csv'
#> 112795 Age a double Bomb C14 '/Users/bocinsky/git/publications/p3k14c/../p3k14c-data/p3k14c_raw.csv'
#> 112812 Age a double Bomb C14 '/Users/bocinsky/git/publications/p3k14c/../p3k14c-data/p3k14c_raw.csv'
#> ...... ... ........ ........ .......................................................................
#> See problems(...) for more details.
#> Joining, by = "Continent"
#> # A tibble: 7 × 6
#> Continent `N Raw Dates (i… `N Scrubbed Dat… `% Dates Retain… `Mean Error (ye…
#> <chr> <int> <int> <dbl> <dbl>
#> 1 Africa 14860 11101 74.7 97.8
#> 2 Asia 21828 13995 64.1 135.
#> 3 Australia 3661 3657 99.9 82.5
#> 4 Central America 1223 1217 99.5 52.9
#> 5 Europe 119106 77144 64.8 97.0
#> 6 North America 102288 64905 63.5 72.0
#> 7 South America 9568 7670 80.2 81.4
#> # … with 1 more variable: Median Error (years) <dbl>
# World map with radiocarbon dates
ggplot() +
geom_sf(data = p3k14c::world) +
geom_sf(data = p3k14c_data,
mapping = aes(color = Continent,
fill = Continent),
alpha = 0.2,
size = 0.75,
shape = 19,
stroke = 0) +
geom_sf_text(data =
tibble::tibble(
x = c(-180,-180,-180),
y = c(-50,0,50),
label = c("50ºS", "0º","50ºN")
) %>%
sf::st_as_sf(coords = c("x","y"),
crs = 4326),
mapping = aes(label = label),
size = 3,
colour = "grey30",
hjust = 1.5) +
scale_x_continuous(expand = ggplot2::expansion(0.05,0)) +
scale_y_continuous(expand = ggplot2::expansion(0,0)) +
ggplot2::labs(x = NULL, y = NULL) +
theme_map(legend.position = c(0.1,0.5)) +
guides(colour = guide_legend(override.aes = list(size = 3,
alpha = 1)))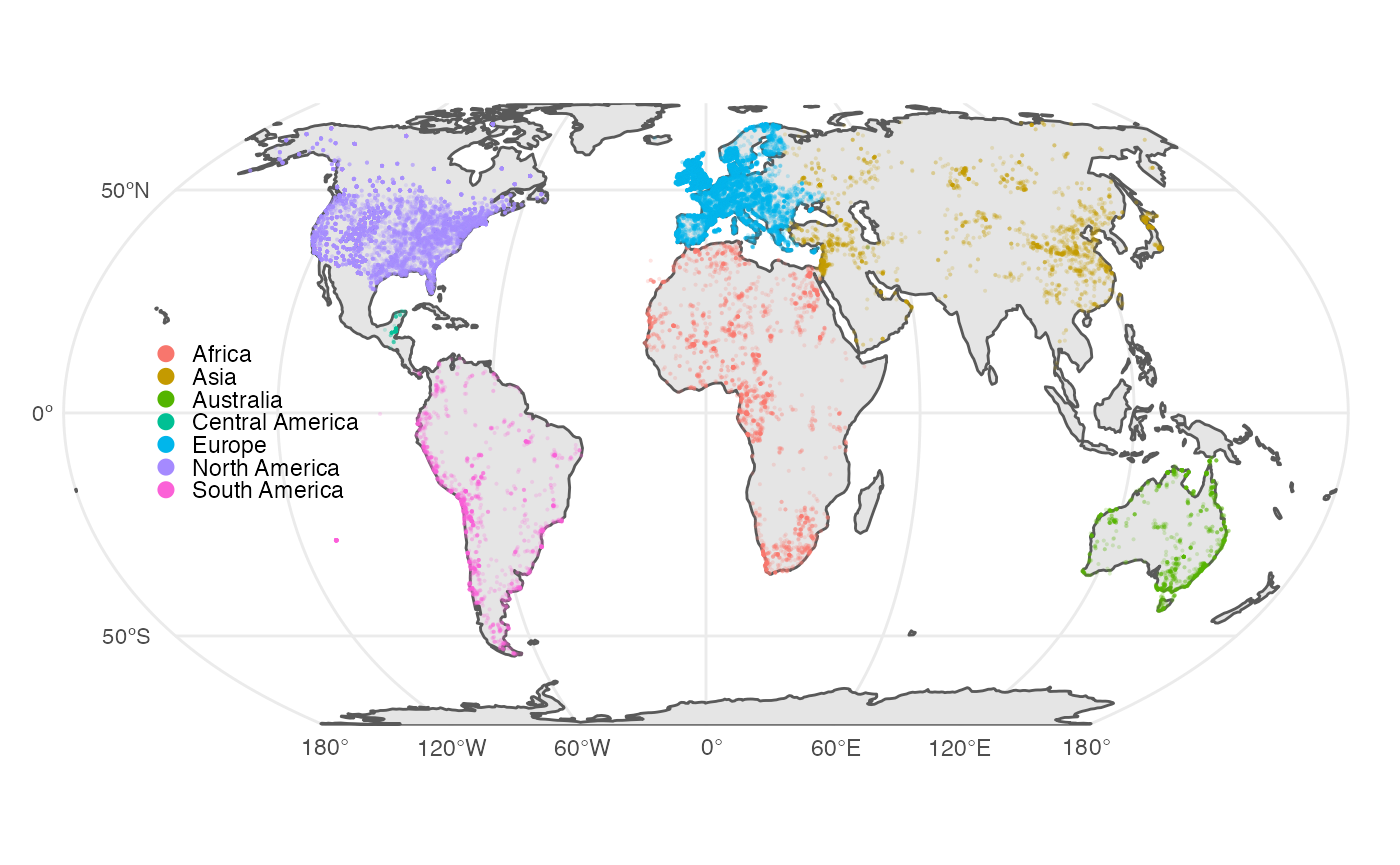
world_plot_ratio <-
p3k14c::world %>%
sf::st_bbox() %>%
as.list() %$%
{(ymax - ymin) / (xmax - xmin)}
fig_width <- 7
fig_height <- fig_width * world_plot_ratio
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure3_global_map.pdf"),
width = fig_width,
height = fig_height,
units = "in"
)
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure3_global_map.png"),
width = fig_width,
height = fig_height,
units = "in"
)
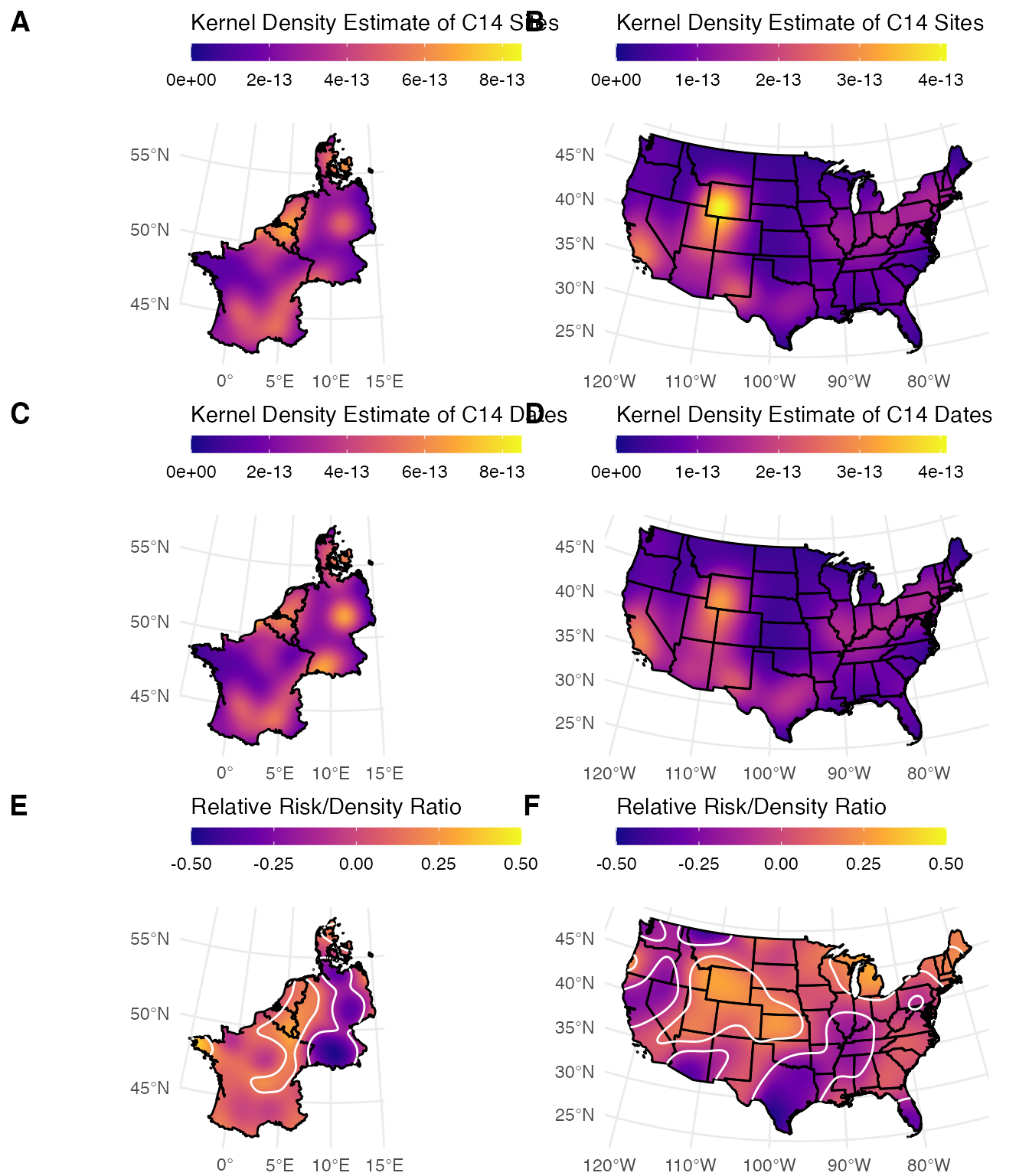
# Spatial windows for risk analysis
windows <-
list(
# Northwestern Europe in Albers Equal Area Conic (ESRI 102013)
`Northwestern Europe` = p3k14c::nw_europe,
# Contiguous USA in Albers Equal Area Conic (ESRI 102003)
`United States` = p3k14c::conus
)
# A function to calculate KDEs and risk surface
as_risk_ppp <-
function(x, w, ...){
x %<>%
# Transform to window projection
sf::st_transform(sf::st_crs(w)) %>%
# Cast to a Spatial Points object
sf::as_Spatial() %>%
# Cast to a Point Pattern object
maptools::as.ppp.SpatialPointsDataFrame() %>%
# Window to extend KDE around globe
window_ppp(
w %>%
sf::st_bbox() %>%
sf::st_as_sfc() %>%
sf::as_Spatial() %>%
maptools::as.owin.SpatialPolygons()
)
# Oversmoothing (OS) bandwidth selector
h0 <-
sparr::OS(x, nstar="npoints")
# Create a KDE of the sites
sites <-
sparr::bivariate.density(pp = x,
h0 = h0,
adapt = FALSE,
edge = "none",
...)
# Create a KDE of the sites,
# weighted by the number of dates at each site
dates <-
sparr::bivariate.density(pp = x,
h0 = h0,
adapt = FALSE,
edge = "none",
weights = x$marks$Dates,
...)
# Calculate the spatial relative risk/density ratio
risk <-
sparr::risk(sites,
dates,
tolerate = TRUE,
...)
# Get two-sided tolerance values
risk$P$v <-
risk$P %>%
as.matrix() %>%
{ 2 * pmin(., 1 - .) }
tibble::lst(
window = w,
sites,
dates,
risk
)
}
# A function to handle plotting
plot_risk <-
function(x){
x$sites %<>%
as_raster.bivden(crs = as(sf::st_crs(x$window), "CRS"),
crop = x$window)
x$dates %<>%
as_raster.bivden(crs = as(sf::st_crs(x$window), "CRS"),
crop = x$window)
x$risk %<>%
as_raster.rrs(crs = as(sf::st_crs(x$window), "CRS"),
crop = x$window)
kde_range = c(0, max(c(x$sites$layer, x$dates$layer), na.rm = TRUE))
x[c("sites","dates")] %<>%
purrr::imap(
~(
ggplot() +
geom_tile(data = .x,
mapping = aes(x = x,
y = y,
fill = layer)) +
geom_sf(data = x$window,
fill = "transparent",
color = "black") +
scale_fill_viridis_c(option = "C",
name = paste0("Kernel Density Estimate of C14 ",
stringr::str_to_title(.y)),
guide = guide_colourbar(
title.position="top",
title.hjust = 0
),
limits = kde_range,
labels = scales::scientific) +
theme_map() +
theme(legend.position = c(0,1),
legend.justification = c(0, 0),
legend.direction = "horizontal",
legend.title = element_text(),
legend.key.width = unit(0.075, 'npc'),
legend.key.height = unit(0.02, 'npc'))
)
)
x[c("risk")] %<>%
purrr::map(
~(
ggplot() +
geom_tile(data = .x,
mapping = aes(x = x,
y = y,
fill = rr)) +
geom_sf(data = x$window,
fill = "transparent",
color = "black") +
geom_contour(data = .x,
mapping = aes(x = x,
y = y,
z = p),
breaks = 0.05,
colour = "white") +
scale_fill_viridis_c(option = "C",
name = "Relative Risk/Density Ratio",
limits = c(-0.5,0.5),
guide = guide_colourbar(
title.position="top",
title.hjust = 0
)) +
theme_map() +
theme(legend.position = c(0,1),
legend.justification = c(0, 0),
legend.direction = "horizontal",
legend.title = element_text(),
legend.key.width = unit(0.075, 'npc'),
legend.key.height = unit(0.02, 'npc'))
)
)
x[c("sites","dates","risk")]
}
# Generate the plots
p3k14c_plots <-
windows %>%
purrr::map(~as_risk_ppp(x = p3k14c_data,
w = .x,
resolution = 1000)) %>%
purrr::map(plot_risk)
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Registered S3 method overwritten by 'spatstat.geom':
#> method from
#> print.boxx cli
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Calculating tolerance contours...
#> Done.
#> Calculating tolerance contours...Done.
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
#> Warning in showSRID(SRS_string, format = "PROJ", multiline = "NO", prefer_proj =
#> prefer_proj): Discarded datum European Datum 1950 in Proj4 definition
ggpubr::ggarrange(
plotlist =
p3k14c_plots %>%
purrr::transpose() %>%
unlist(recursive = FALSE),
ncol = 2,
nrow = 3,
align = "none",
labels =
c("A","B","C","D","E","F"),
legend = "top"
)
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure4_risk.pdf"),
width = 7,
height = 8,
units = "in"
)
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure4_risk.png"),
width = 7,
height = 8,
units = "in"
)
# Regions for quality assessment
regional_dates_sites <-
list(
`China (provinces)` =
# Data from Hosner et al. 2016,
# which provides site count by province for China
readr::read_tsv("https://doi.pangaea.de/10.1594/PANGAEA.860072?format=textfile",
skip = 20) %>%
dplyr::select(
Province = Volume
) %>%
dplyr::mutate(Province = factor(Province),
Province = forcats::fct_recode(Province,
Xizang = "Tibet Autonomous Region",
`Inner Mongol` = "Inner Mongolia Autonomous Region",
Ningxia = "Ningxia Hui Autonomous Region",
Xinjiang = "Xinjiang Uyghur Autonomous Region"),
Province = as.character(Province)) %>%
dplyr::group_by(Province) %>%
dplyr::count(name = "Recorded Sites") %>%
dplyr::left_join(
p3k14c::p3k14c_data %>%
dplyr::filter(Country == "China") %>%
dplyr::group_by(Province, Long,Lat, SiteID, SiteName) %>%
dplyr::count(name = "Dates") %>%
dplyr::group_by(Province) %>%
dplyr::summarise(`Dated Sites` = dplyr::n(),
Dates = sum(Dates))
) %>%
dplyr::ungroup() %>%
dplyr::left_join(
rnaturalearth::ne_states(country = "China",
returnclass = "sf") %>%
dplyr::select(Province = name)
) %>%
dplyr::arrange(Province) %>%
sf::st_as_sf() %>%
sf::st_transform("+proj=aea +lat_1=27 +lat_2=45 +lat_0=35 +lon_0=105 +x_0=0 +y_0=0 +ellps=WGS84 +datum=WGS84 +units=m +no_defs") %>%
dplyr::mutate(Area = sf::st_area(geometry) %>%
units::set_units("km^2"),
`Recorded Sites density` = units::drop_units(`Recorded Sites`/Area) * 10000,
`Dated Sites density` = units::drop_units(`Dated Sites`/Area) * 10000,
`Dates density` = units::drop_units(Dates/Area) * 10000) %>%
dplyr::rename(`Study Unit` = Province)
,
`Western Africa (countries)` =
# Data from Kay et al. 2019,
# which provides site data by country for Western Africa
here::here("inst/western_africa_sites.csv") %>%
readr::read_csv() %>%
dplyr::mutate(Country = factor(Country),
Country = forcats::fct_recode(Country,
`Burkina Faso` = "BurkinaFaso",
`Central African Republic` = "CAR",
`Comoros Islands` = "ComorosIslands",
`Democratic Republic of the Congo` = "DRC",
`Equatorial Guinea` = "Equat.Guinea",
`Gran Canaria` = "GranCanaria,Spain",
`Sierra Leone` = "SierraLeone",
`South Africa` = "SouthAfrica",
`Western Sahara` = "WesternSahara"),
Country = as.character(Country)) %>%
dplyr::filter(
Country %in% c("Benin",
"Burkina Faso",
"Cameroon",
"Chad",
"Congo",
"Democratic Republic of the Congo",
"Equatorial Guinea",
"Gabon",
"Ghana",
"Guinea",
"IvoryCoast",
"Liberia",
"Mali",
"Mauritania",
"Niger",
"Nigeria",
"Togo")) %>%
dplyr::group_by(Country) %>%
dplyr::count(name = "Recorded Sites") %>%
dplyr::left_join(
p3k14c::p3k14c_data %>%
dplyr::group_by(Country, Long, Lat, SiteID, SiteName) %>%
dplyr::count(name = "Dates") %>%
dplyr::group_by(Country) %>%
dplyr::summarise(`Dated Sites` = dplyr::n(),
Dates = sum(Dates))
) %>%
dplyr::ungroup() %>%
dplyr::left_join(
rnaturalearth::ne_countries(continent = "africa",
returnclass = "sf") %>%
dplyr::select(Country = admin) %>%
dplyr::mutate(Country = ifelse(Country == "Ivory Coast",
"IvoryCoast",
Country),
Country = ifelse(Country == "Republic of Congo",
"Congo",
Country)
)
) %>%
dplyr::arrange(Country) %>%
sf::st_as_sf() %>%
sf::st_transform("ESRI:102022") %>%
dplyr::mutate(Area = sf::st_area(geometry) %>%
units::set_units("km^2"),
`Recorded Sites density` = units::drop_units(`Recorded Sites`/Area) * 10000,
`Dated Sites density` = units::drop_units(`Dated Sites`/Area) * 10000,
`Dates density` = units::drop_units(Dates/Area) * 10000) %>%
dplyr::rename(`Study Unit` = Country)
) %>%
purrr::map(sf::st_drop_geometry) %>%
dplyr::bind_rows(.id = "Region") %>%
dplyr::select(Region,
`Study Unit`,
`Recorded Sites density`,
`Dated Sites density`,
`Dates density`) %>%
dplyr::rename(Recorded = `Recorded Sites density`,
Dated = `Dated Sites density`) %>%
tidyr::pivot_longer(Recorded:Dated,
names_to = "Site Type",
values_to = "Sites density") %>%
dplyr::mutate(Region = factor(Region,
levels = c("China (provinces)",
"Western Africa (countries)"),
labels = c(
"China (provinces)",
"W. Africa (countries)"
),
ordered = TRUE),
`Site Type` = factor(`Site Type`,
levels = c("Recorded",
"Dated"),
labels = c("Recorded Sites",
"Dated Sites"),
ordered = TRUE))
#>
#> ── Column specification ────────────────────────────────────────────────────────
#> cols(
#> Site = col_character(),
#> Volume = col_character(),
#> `Page(s)` = col_character(),
#> Type = col_character(),
#> Area = col_character(),
#> `Cult age` = col_character(),
#> `Age max [ka]` = col_double(),
#> `Age dated min [ka]` = col_double(),
#> `Age [ka BP]` = col_double(),
#> Longitude = col_double(),
#> Latitude = col_double()
#> )
#> Joining, by = "Province"
#> Joining, by = "Province"
#>
#> ── Column specification ────────────────────────────────────────────────────────
#> cols(
#> Site_ID = col_double(),
#> Site_Name = col_character(),
#> Country = col_character()
#> )
#> Joining, by = "Country"
#> Joining, by = "Country"
regional_dates_sites_sma <-
regional_dates_sites %>%
dplyr::group_by(Region, `Site Type`) %>%
dplyr::summarise(
sma = list({
smatr::sma(log10(`Dates density`) ~ log10(`Sites density`)) %$%
tibble::tibble(elevation = coef[[1]]$`coef(SMA)`[[1]],
slope = coef[[1]]$`coef(SMA)`[[2]],
p = pval[[1]])
})
) %>%
tidyr::unnest(sma) %>%
dplyr::ungroup() %>%
dplyr::mutate(panel = c("A", "B", "C", "D"))
#> `summarise()` has grouped output by 'Region'. You can override using the `.groups` argument.
regional_dates_sites %>%
# dplyr::rename(Site_Type = `Site Type`) %>%
ggplot(aes(x = `Sites density`,
y = `Dates density`)) +
geom_point() +
geom_abline(
data = regional_dates_sites_sma,
aes(
intercept = elevation,
slope = slope
),
color = "red"
) +
geom_text(
data = regional_dates_sites_sma,
aes(
x = 0,
y = Inf,
label = paste0("m = ", round(slope, 3),"\n",
ifelse(p < 0.001, "P < 0.001",
paste("p =", round(p, 3))
)
)),
hjust = -0.1,
vjust = 2.5,
color = "red"
) +
geom_text(
data = regional_dates_sites_sma,
aes(
x = 0,
y = Inf,
label = panel
),
hjust = -0.1,
vjust = 1.1,
size = 10,
fontface = "bold"
) +
scale_x_log10() +
scale_y_log10() +
labs(
x = expression("Sites per 10,000" ~ km^2),
y = expression("Dates per 10,000" ~ km^2)
) +
theme_map(strip.text = element_text(size = 12,
face = "bold",
hjust = 0.5)) +
theme(axis.title = ggplot2::element_text(),
plot.margin = ggplot2::margin(t = 1, r = 5, b = 1, l = 1, unit = "mm")) +
facet_grid(Region ~ `Site Type`)
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Removed 4 rows containing missing values (geom_point).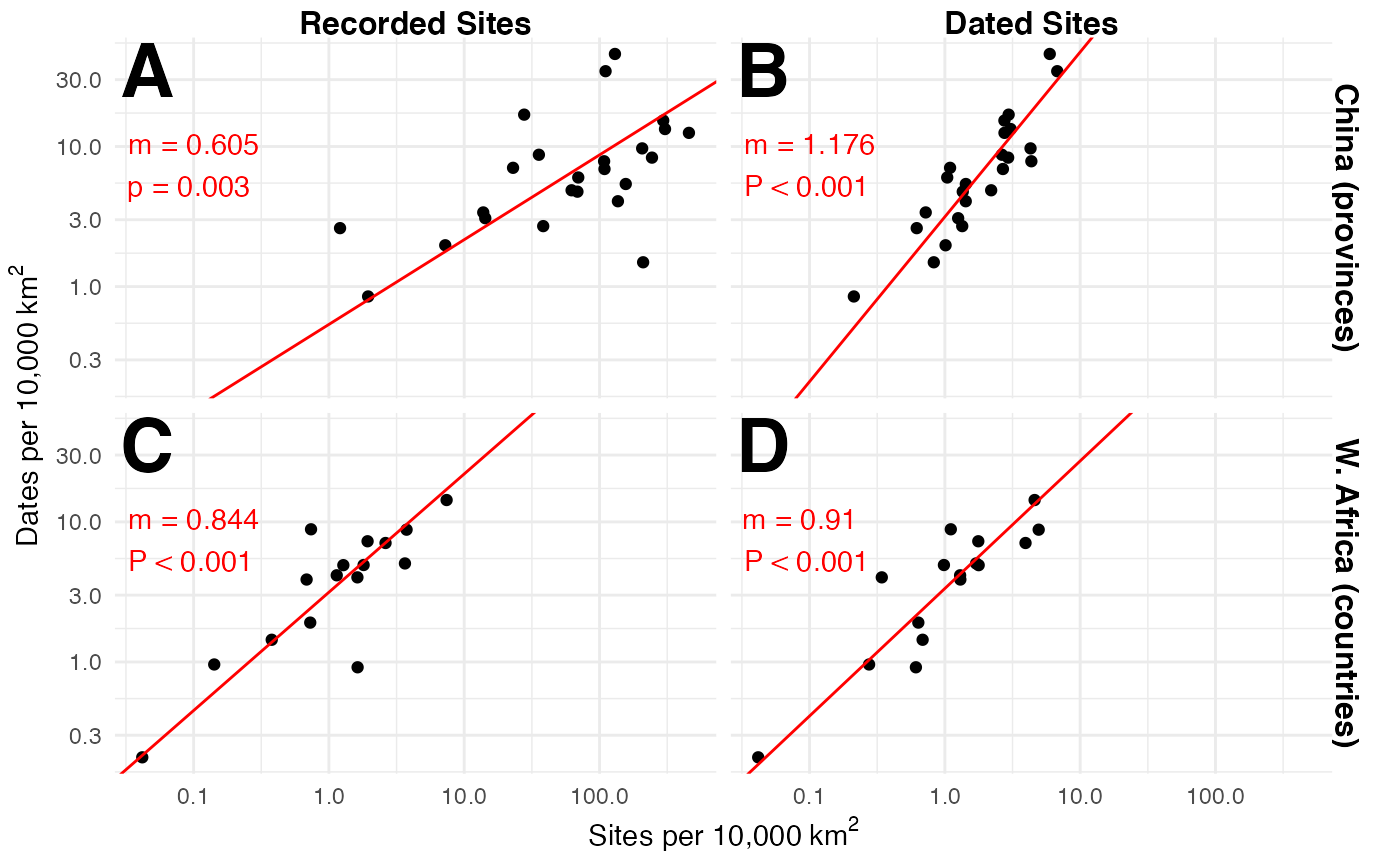
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure5_regional_quality_assessment.pdf"),
width = 7,
height = 9/2,
units = "in"
)
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Removed 4 rows containing missing values (geom_point).
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure5_regional_quality_assessment.png"),
width = 7,
height = 9/2,
units = "in"
)
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Removed 4 rows containing missing values (geom_point).
# NaturalEarth dataset of admin 1 boundaries
sf::sf_use_s2(FALSE)
#> Spherical geometry (s2) switched off
admin_1 <-
rnaturalearth::ne_states(returnclass = "sf") %>%
dplyr::select(`Admin1` = name) %>%
sf::st_make_valid() %>%
sf::st_cast("MULTIPOLYGON") %>%
sf::st_transform(4326)
# sf::sf_use_s2(TRUE)
# Risk-type global assessment by continent
global_dates_sites <-
p3k14c::p3k14c_data %>%
dplyr::group_by(Continent, Country, Long, Lat, SiteID, SiteName) %>%
dplyr::count(name = "Dates") %>%
tidyr::drop_na(Long, Lat) %>%
sf::st_as_sf(
coords = c("Long", "Lat"),
crs = "EPSG:4326",
remove = FALSE
) %>%
sf::st_intersection(admin_1) %>%
sf::st_drop_geometry() %>%
dplyr::group_by(Continent, `Admin1`) %>%
dplyr::summarise(`Dated Sites` = dplyr::n(),
Dates = sum(Dates)) %>%
dplyr::left_join(admin_1) %>%
dplyr::arrange(Continent, `Admin1`) %>%
sf::st_as_sf() %>%
sf::st_transform("EPSG:8857") %>%
dplyr::mutate(Area = sf::st_area(geometry) %>%
units::set_units("km^2"),
`Dated Sites density` = units::drop_units(`Dated Sites`/Area) * 10000,
`Dates density` = units::drop_units(Dates/Area) * 10000) %>%
dplyr::ungroup() %>%
dplyr::mutate(Continent = factor(Continent,
levels = c("North America",
"Central America",
"South America",
"Europe",
"Africa",
"Asia",
"Australia"),
ordered = TRUE))
#> although coordinates are longitude/latitude, st_intersection assumes that they are planar
#> Warning: attribute variables are assumed to be spatially constant throughout all
#> geometries
#> `summarise()` has grouped output by 'Continent'. You can override using the `.groups` argument.
#> Joining, by = "Admin1"
global_dates_sites_sma <-
global_dates_sites %>%
sf::st_drop_geometry() %>%
dplyr::group_by(Continent) %>%
dplyr::summarise(
sma = list({
smatr::sma(log10(`Dates density`) ~ log10(`Dated Sites density`)) %$%
tibble::tibble(elevation = coef[[1]]$`coef(SMA)`[[1]],
slope = coef[[1]]$`coef(SMA)`[[2]],
p = pval[[1]])
})
) %>%
tidyr::unnest(sma) %>%
dplyr::ungroup() %>%
dplyr::mutate(panel = c("A", "B", "C", "D", "E", "F", "G"))
global_dates_sites %>%
ggplot(aes(x = `Dated Sites density`,
y = `Dates density`)) +
geom_point() +
geom_abline(
data = global_dates_sites_sma,
aes(
intercept = elevation,
slope = slope
),
color = "red"
) +
geom_text(
data = global_dates_sites_sma,
aes(
x = 0,
y = Inf,
label = paste0("m = ", round(slope, 3),"\n",
ifelse(p < 0.001, "P < 0.001",
paste("p =", round(p, 3))
)
)),
hjust = -0.1,
vjust = 2.5,
color = "red"
) +
geom_text(
data = global_dates_sites_sma,
aes(
x = 0,
y = Inf,
label = panel
),
hjust = -0.1,
vjust = 1.1,
size = 10,
fontface = "bold"
) +
scale_x_log10() +
scale_y_log10() +
labs(
x = expression("Dated Sites per 10,000" ~ km^2),
y = expression("Dates per 10,000" ~ km^2)
) +
theme_map(strip.text = element_text(size = 12,
face = "bold",
hjust = 0.5)) +
theme(axis.title = ggplot2::element_text(),
plot.margin = ggplot2::margin(t = 1, r = 5, b = 1, l = 1, unit = "mm")
) +
facet_wrap("Continent",
ncol = 2)
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Transformation introduced infinite values in continuous x-axis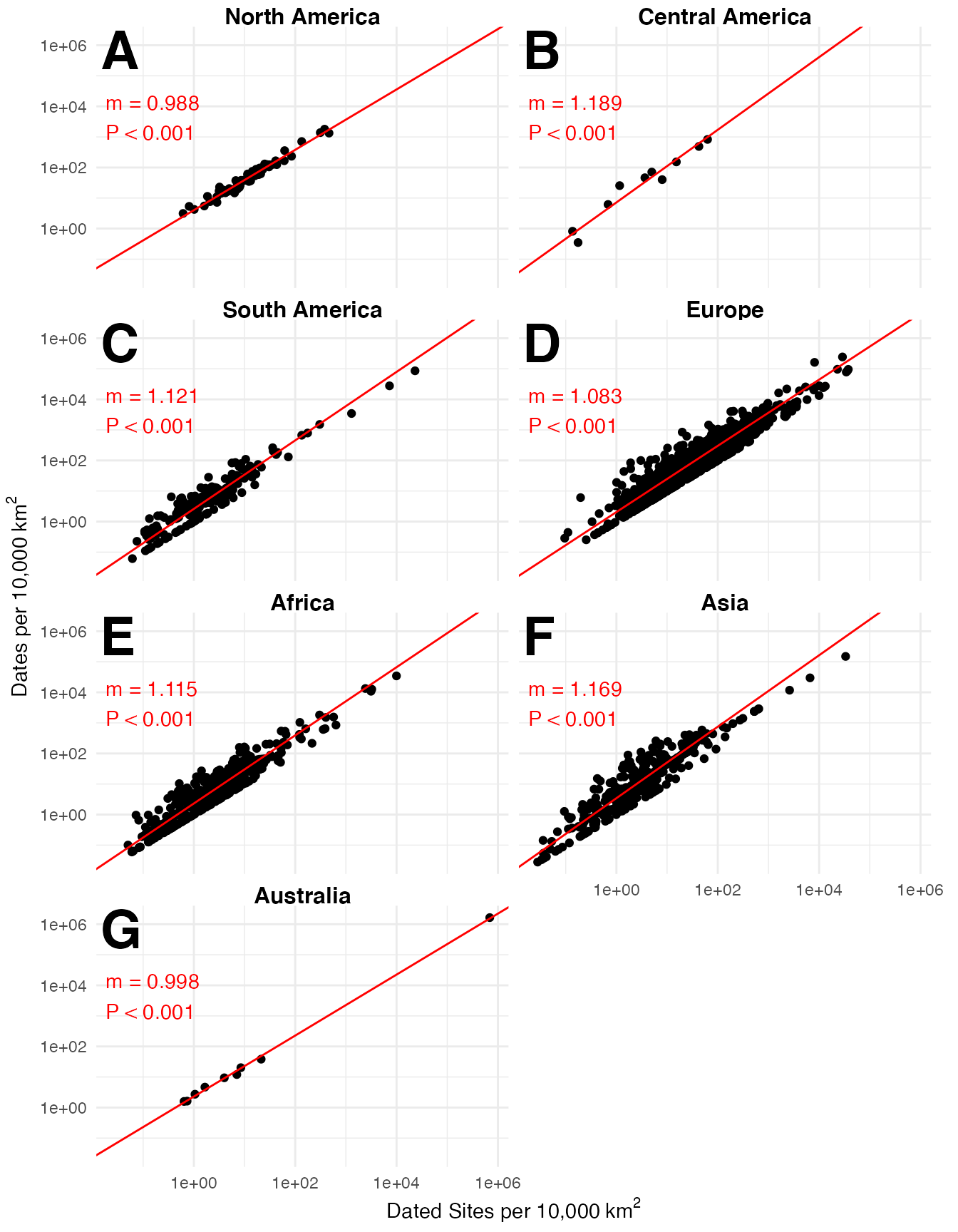
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure6_continent_bias.pdf"),
width = 7,
height = 9,
units = "in"
)
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Transformation introduced infinite values in continuous x-axis
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure6_continent_bias.png"),
width = 7,
height = 9,
units = "in"
)
#> Warning: Transformation introduced infinite values in continuous x-axis
#> Warning: Transformation introduced infinite values in continuous x-axisoptions(tigris_use_cache = FALSE)
conus_counties <-
tigris::counties(year = 2020) %>%
dplyr::select(County = NAME,
STATEFP) %>%
sf::st_transform(crs = "ESRI:102003") %>%
dplyr::left_join(tigris::states(year = 2020) %>%
sf::st_drop_geometry() %>%
dplyr::select(STATEFP, State = NAME)) %>%
dplyr::select(County, State) %>%
tidyr::unite(County:State, col = "County", sep = ", ")
#>
|
| | 0%
|
| | 1%
|
|= | 1%
|
|= | 2%
|
|== | 2%
|
|== | 3%
|
|== | 4%
|
|=== | 4%
|
|=== | 5%
|
|==== | 5%
|
|==== | 6%
|
|===== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 9%
|
|======= | 10%
|
|======= | 11%
|
|======== | 11%
|
|======== | 12%
|
|========= | 13%
|
|========= | 14%
|
|========== | 14%
|
|========== | 15%
|
|=========== | 15%
|
|=========== | 16%
|
|============ | 17%
|
|============= | 18%
|
|============= | 19%
|
|============== | 19%
|
|============== | 20%
|
|============== | 21%
|
|=============== | 21%
|
|=============== | 22%
|
|================ | 22%
|
|================ | 23%
|
|================ | 24%
|
|================= | 24%
|
|================= | 25%
|
|================== | 25%
|
|================== | 26%
|
|=================== | 27%
|
|=================== | 28%
|
|==================== | 28%
|
|==================== | 29%
|
|===================== | 29%
|
|===================== | 30%
|
|===================== | 31%
|
|======================= | 32%
|
|======================= | 33%
|
|======================= | 34%
|
|======================== | 34%
|
|========================= | 36%
|
|========================== | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|============================== | 44%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|=================================== | 50%
|
|=================================== | 51%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 52%
|
|===================================== | 53%
|
|===================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|======================================== | 56%
|
|======================================== | 57%
|
|======================================== | 58%
|
|========================================= | 58%
|
|========================================== | 60%
|
|========================================== | 61%
|
|=========================================== | 61%
|
|=========================================== | 62%
|
|============================================ | 62%
|
|============================================ | 63%
|
|============================================= | 65%
|
|============================================== | 65%
|
|============================================== | 66%
|
|=============================================== | 66%
|
|=============================================== | 67%
|
|=============================================== | 68%
|
|================================================ | 68%
|
|================================================= | 69%
|
|================================================= | 70%
|
|================================================= | 71%
|
|================================================== | 71%
|
|================================================== | 72%
|
|=================================================== | 72%
|
|=================================================== | 73%
|
|==================================================== | 74%
|
|==================================================== | 75%
|
|===================================================== | 75%
|
|===================================================== | 76%
|
|====================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 79%
|
|======================================================== | 79%
|
|======================================================== | 80%
|
|======================================================== | 81%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 84%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 86%
|
|============================================================= | 87%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 89%
|
|=============================================================== | 90%
|
|=============================================================== | 91%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 93%
|
|================================================================= | 94%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|==================================================================== | 98%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%
#>
|
| | 0%
|
| | 1%
|
|= | 1%
|
|=== | 4%
|
|=== | 5%
|
|==== | 6%
|
|===== | 6%
|
|===== | 7%
|
|====== | 8%
|
|====== | 9%
|
|======= | 10%
|
|================== | 25%
|
|===================== | 30%
|
|====================== | 32%
|
|======================= | 32%
|
|======================= | 33%
|
|======================== | 34%
|
|======================== | 35%
|
|========================= | 36%
|
|========================== | 37%
|
|========================== | 38%
|
|=========================== | 38%
|
|=========================== | 39%
|
|============================ | 39%
|
|============================ | 40%
|
|============================ | 41%
|
|============================= | 41%
|
|============================= | 42%
|
|============================== | 42%
|
|============================== | 43%
|
|=============================== | 44%
|
|================================ | 45%
|
|================================ | 46%
|
|================================= | 46%
|
|================================= | 47%
|
|================================= | 48%
|
|================================== | 48%
|
|================================== | 49%
|
|=================================== | 50%
|
|==================================== | 51%
|
|==================================== | 52%
|
|===================================== | 53%
|
|====================================== | 54%
|
|====================================== | 55%
|
|======================================= | 55%
|
|======================================= | 56%
|
|================================================= | 70%
|
|=================================================== | 73%
|
|===================================================== | 76%
|
|====================================================== | 77%
|
|======================================================= | 78%
|
|======================================================= | 79%
|
|======================================================== | 79%
|
|======================================================== | 80%
|
|========================================================= | 81%
|
|========================================================= | 82%
|
|========================================================== | 82%
|
|========================================================== | 83%
|
|=========================================================== | 84%
|
|=========================================================== | 85%
|
|============================================================ | 85%
|
|============================================================ | 86%
|
|============================================================= | 87%
|
|============================================================= | 88%
|
|============================================================== | 88%
|
|============================================================== | 89%
|
|=============================================================== | 90%
|
|================================================================ | 91%
|
|================================================================ | 92%
|
|================================================================= | 92%
|
|================================================================= | 93%
|
|================================================================== | 94%
|
|================================================================== | 95%
|
|=================================================================== | 96%
|
|==================================================================== | 97%
|
|===================================================================== | 98%
|
|===================================================================== | 99%
|
|======================================================================| 99%
|
|======================================================================| 100%
#> Joining, by = "STATEFP"
conus_dates_sites <-
list(
`State` =
# Data from many sources documented in conus_site_counts.csv
here::here("inst/conus_site_counts.csv") %>%
readr::read_csv() %>%
dplyr::select(State,
`Recorded Sites` = nSites) %>%
na.omit() %>%
dplyr::left_join(
p3k14c::p3k14c_data %>%
dplyr::filter(Country == "USA") %>%
dplyr::group_by(Province, Long, Lat, SiteID, SiteName) %>%
dplyr::count(name = "Dates") %>%
dplyr::group_by(State = Province) %>%
dplyr::summarise(`Dated Sites` = dplyr::n(),
Dates = sum(Dates))
) %>%
dplyr::left_join(
rnaturalearth::ne_states(country = "United States of America",
returnclass = "sf") %>%
dplyr::select(State = name)
) %>%
dplyr::arrange(State) %>%
sf::st_as_sf() %>%
sf::st_transform("ESRI:102003") %>%
dplyr::mutate(Area = sf::st_area(geometry) %>%
units::set_units("km^2"),
`Recorded Sites density` = units::drop_units(`Recorded Sites`/Area) * 10000,
`Dated Sites density` = units::drop_units(`Dated Sites`/Area) * 10000,
`Dates density` = units::drop_units(Dates/Area) * 10000) %>%
sf::st_drop_geometry() %>%
dplyr::select(Name = State,
`Recorded Sites density`,
`Dated Sites density`,
`Dates density`) %>%
dplyr::rename(Recorded = `Recorded Sites density`,
Dated = `Dated Sites density`) %>%
tidyr::pivot_longer(Recorded:Dated,
names_to = "Site Type",
values_to = "Sites density"),
`County` =
p3k14c::p3k14c_data %>%
dplyr::filter(Country == "USA") %>%
dplyr::group_by(Province, Long, Lat, SiteID, SiteName) %>%
dplyr::count(name = "Dates") %>%
tidyr::drop_na(Long, Lat) %>%
sf::st_as_sf(coords = c("Long", "Lat"),
crs = "EPSG:4326") %>%
sf::st_transform("ESRI:102003") %>%
sf::st_intersection(
conus_counties
) %>%
sf::st_drop_geometry() %>%
dplyr::group_by(County) %>%
dplyr::summarise(`Dated Sites` = dplyr::n(),
Dates = sum(Dates)) %>%
dplyr::left_join(
conus_counties
) %>%
dplyr::arrange(County) %>%
sf::st_as_sf() %>%
dplyr::mutate(Area = sf::st_area(geometry) %>%
units::set_units("km^2"),
`Dated Sites density` = units::drop_units(`Dated Sites`/Area) * 10000,
`Dates density` = units::drop_units(Dates/Area) * 10000) %>%
sf::st_drop_geometry() %>%
dplyr::select(Name = County,
`Dated Sites density`,
`Dates density`) %>%
dplyr::rename(Dated = `Dated Sites density`) %>%
tidyr::pivot_longer(Dated,
names_to = "Site Type",
values_to = "Sites density")
) %>%
dplyr::bind_rows(.id = "Scale") %>%
dplyr::mutate(Scale = factor(Scale,
levels = c("State",
"County"),
ordered = TRUE),
`Site Type` = factor(`Site Type`,
levels = c("Recorded",
"Dated"),
ordered = TRUE)
) %>%
dplyr::arrange(dplyr::desc(`Dates density`)) %>%
dplyr::mutate(group = paste0(`Site Type`, " Sites by ", Scale))
#>
#> ── Column specification ────────────────────────────────────────────────────────
#> cols(
#> State = col_character(),
#> nSites = col_double(),
#> precision = col_character(),
#> `Site # Source` = col_character(),
#> `Email/Weblink` = col_character(),
#> Conducted = col_character(),
#> Status = col_character(),
#> Comment = col_character()
#> )
#> Joining, by = "State"
#> Joining, by = "State"
#> Warning: attribute variables are assumed to be spatially constant throughout all
#> geometries
#> Joining, by = "County"
conus_dates_sites_sma <-
conus_dates_sites %>%
dplyr::group_by(group) %>%
dplyr::summarise(
sma = list({
smatr::sma(`Dates density` ~ `Sites density`) %$%
tibble::tibble(elevation = coef[[1]]$`coef(SMA)`[[1]],
slope = coef[[1]]$`coef(SMA)`[[2]],
p = pval[[1]])
})
) %>%
tidyr::unnest(sma) %>%
dplyr::ungroup() %>%
dplyr::mutate(panel = c("A", "C", "E"))
conus_dates_sites_sma_log <-
conus_dates_sites %>%
dplyr::group_by(group) %>%
dplyr::summarise(
sma = list({
smatr::sma(log10(`Dates density`) ~ log10(`Sites density`)) %$%
tibble::tibble(elevation = coef[[1]]$`coef(SMA)`[[1]],
slope = coef[[1]]$`coef(SMA)`[[2]],
p = pval[[1]])
})
) %>%
tidyr::unnest(sma) %>%
dplyr::ungroup() %>%
dplyr::mutate(panel = c("B", "D", "F"))
conus_no_log <-
conus_dates_sites %>%
ggplot(aes(x = `Sites density`,
y = `Dates density`)) +
geom_point() +
geom_abline(
data = conus_dates_sites_sma,
aes(
intercept = elevation,
slope = slope
),
color = "red"
) +
geom_text(
data = conus_dates_sites_sma,
aes(
x = 0,
y = Inf,
label = paste0("m = ", round(slope, 3),"\n",
ifelse(p < 0.001, "P < 0.001",
paste("p =", round(p, 3))
)
)),
hjust = -0.1,
vjust = 2.5,
color = "red"
) +
geom_text(
data = conus_dates_sites_sma,
aes(
x = 0,
y = Inf,
label = panel
),
hjust = -0.1,
vjust = 1.1,
size = 10,
fontface = "bold"
) +
scale_x_continuous(limits = c(0,NA),
expand = expansion(c(0,0.05),0)) +
scale_y_continuous(limits = c(0,NA),
expand = expansion(c(0,0.05),0)) +
labs(
x = expression("Sites per 10,000" ~ km^2),
y = expression("Dates per 10,000" ~ km^2)
) +
theme_map(strip.text = element_text(size = 12,
face = "bold",
hjust = 0.5)) +
theme(axis.title = ggplot2::element_text(),
plot.margin = ggplot2::margin(t = 1, r = 1, b = 1, l = 1, unit = "mm")
) +
facet_wrap(~ group,
scales = "free",
ncol = 1)
conus_log <-
conus_dates_sites %>%
ggplot(aes(x = `Sites density`,
y = `Dates density`)) +
geom_point() +
geom_abline(
data = conus_dates_sites_sma_log,
aes(
intercept = elevation,
slope = slope
),
color = "red"
) +
geom_text(
data = conus_dates_sites_sma_log,
aes(
x = 1,
y = Inf,
label = paste0("m = ", round(slope, 3),"\n",
ifelse(p < 0.001, "P < 0.001",
paste("p =", round(p, 3))
)
)),
hjust = -0.1,
vjust = 2.5,
color = "red"
) +
geom_text(
data = conus_dates_sites_sma_log,
aes(
x = 1,
y = Inf,
label = panel
),
hjust = -0.1,
vjust = 1.1,
size = 10,
fontface = "bold"
) +
scale_x_log10(limits = c(1,10000),
expand = expansion(c(0,0.05),0)) +
scale_y_log10(limits = c(1,100000),
expand = expansion(c(0,0.05),0)) +
labs(
x = expression("Sites per 10,000" ~ km^2),
y = NULL
) +
theme_map(strip.text = element_text(size = 12,
face = "bold",
hjust = 0.5)) +
theme(axis.title = ggplot2::element_text(),
plot.margin = ggplot2::margin(t = 1, r = 5, b = 1, l = 1, unit = "mm")
) +
facet_wrap(~ group,
ncol = 1)
ggpubr::ggarrange(
conus_no_log,
conus_log,
ncol = 2,
align = "hv"
)
#> Warning: Removed 8 rows containing missing values (geom_point).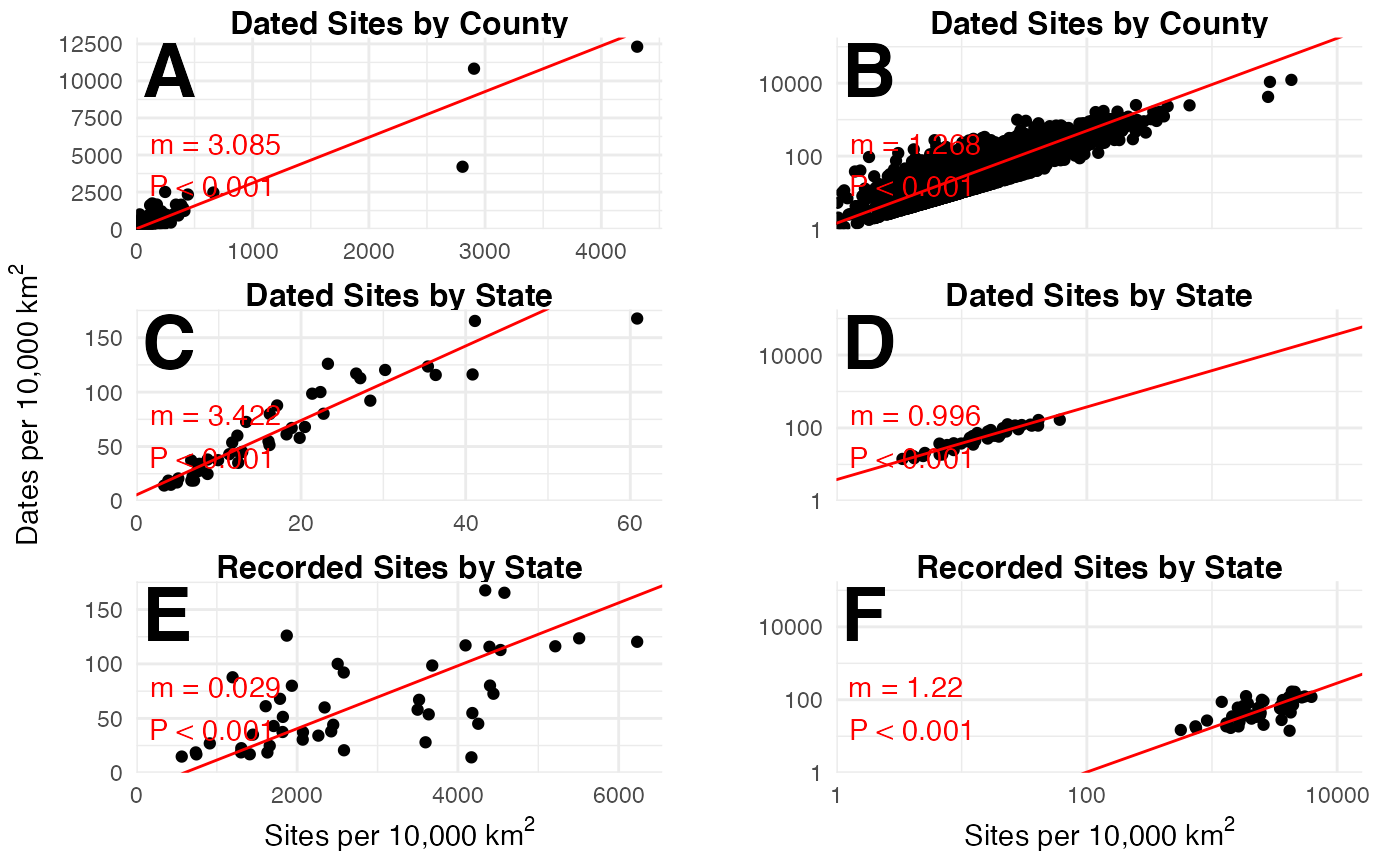
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure7_conus_bias.pdf"),
width = 7,
height = 9 * (3/4),
units = "in"
)
ggplot2::ggsave(
filename = here::here("vignettes/articles/figures/Figure7_conus_bias.png"),
width = 7,
height = 9 * (3/4),
units = "in"
)References
Colophon
This report was generated on 2021-08-02 21:25:19 using the following computational environment and dependencies:
# which R packages and versions?
if ("devtools" %in% installed.packages()) devtools::session_info()
#> ─ Session info ───────────────────────────────────────────────────────────────
#> setting value
#> version R version 4.1.0 (2021-05-18)
#> os macOS Big Sur 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz America/Denver
#> date 2021-08-02
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date lib
#> abind 1.4-5 2016-07-21 [2]
#> assertthat 0.2.1 2019-03-21 [2]
#> backports 1.2.1 2020-12-09 [2]
#> broom 0.7.8 2021-06-24 [2]
#> bslib 0.2.5.1 2021-05-18 [2]
#> cachem 1.0.5 2021-05-15 [2]
#> callr 3.7.0 2021-04-20 [2]
#> car 3.0-11 2021-06-27 [2]
#> carData 3.0-4 2020-05-22 [2]
#> cellranger 1.1.0 2016-07-27 [2]
#> class 7.3-19 2021-05-03 [2]
#> classInt 0.4-3 2020-04-07 [2]
#> cli 3.0.1 2021-07-17 [2]
#> codetools 0.2-18 2020-11-04 [2]
#> colorspace 2.0-2 2021-06-24 [2]
#> cowplot 1.1.1 2020-12-30 [2]
#> crayon 1.4.1 2021-02-08 [2]
#> curl 4.3.2 2021-06-23 [2]
#> data.table 1.14.0 2021-02-21 [2]
#> DBI 1.1.1 2021-01-15 [2]
#> deldir 0.2-10 2021-02-16 [2]
#> desc 1.3.0 2021-03-05 [2]
#> devtools 2.4.2 2021-06-07 [2]
#> digest 0.6.27 2020-10-24 [2]
#> doParallel 1.0.16 2020-10-16 [2]
#> dplyr 1.0.7 2021-06-18 [2]
#> e1071 1.7-7 2021-05-23 [2]
#> ellipsis 0.3.2 2021-04-29 [2]
#> evaluate 0.14 2019-05-28 [2]
#> fansi 0.5.0 2021-05-25 [2]
#> farver 2.1.0 2021-02-28 [2]
#> fastmap 1.1.0 2021-01-25 [2]
#> forcats 0.5.1 2021-01-27 [2]
#> foreach 1.5.1 2020-10-15 [2]
#> foreign 0.8-81 2020-12-22 [2]
#> fs 1.5.0 2020-07-31 [2]
#> generics 0.1.0 2020-10-31 [2]
#> ggplot2 * 3.3.5 2021-06-25 [2]
#> ggpubr 0.4.0 2020-06-27 [2]
#> ggsignif 0.6.2 2021-06-14 [2]
#> glue 1.4.2 2020-08-27 [2]
#> goftest 1.2-2 2019-12-02 [2]
#> gtable 0.3.0 2019-03-25 [2]
#> haven 2.4.1 2021-04-23 [2]
#> here 1.0.1 2020-12-13 [2]
#> highr 0.9 2021-04-16 [2]
#> hms 1.1.0 2021-05-17 [2]
#> htmltools 0.5.1.1 2021-01-22 [2]
#> httr 1.4.2 2020-07-20 [2]
#> isoband 0.2.5 2021-07-13 [2]
#> iterators 1.0.13 2020-10-15 [2]
#> jquerylib 0.1.4 2021-04-26 [2]
#> jsonlite 1.7.2 2020-12-09 [2]
#> KernSmooth 2.23-20 2021-05-03 [2]
#> knitr 1.33 2021-04-24 [2]
#> labeling 0.4.2 2020-10-20 [2]
#> lattice 0.20-44 2021-05-02 [2]
#> lifecycle 1.0.0 2021-02-15 [2]
#> magrittr * 2.0.1 2020-11-17 [2]
#> maptools 1.1-1 2021-03-15 [2]
#> Matrix 1.3-3 2021-05-04 [2]
#> memoise 2.0.0 2021-01-26 [2]
#> mgcv 1.8-36 2021-06-01 [2]
#> misc3d 0.9-0 2020-09-06 [2]
#> munsell 0.5.0 2018-06-12 [2]
#> nlme 3.1-152 2021-02-04 [2]
#> openxlsx 4.2.4 2021-06-16 [2]
#> p3k14c * 1.0.0 2021-08-03 [1]
#> pillar 1.6.2 2021-07-29 [2]
#> pkgbuild 1.2.0 2020-12-15 [2]
#> pkgconfig 2.0.3 2019-09-22 [2]
#> pkgdown 1.6.1 2020-09-12 [2]
#> pkgload 1.2.1 2021-04-06 [2]
#> polyclip 1.10-0 2019-03-14 [2]
#> prettyunits 1.1.1 2020-01-24 [2]
#> processx 3.5.2 2021-04-30 [2]
#> proxy 0.4-26 2021-06-07 [2]
#> ps 1.6.0 2021-02-28 [2]
#> purrr 0.3.4 2020-04-17 [2]
#> R6 2.5.0 2020-10-28 [2]
#> ragg 1.1.3 2021-06-09 [2]
#> rappdirs 0.3.3 2021-01-31 [2]
#> raster 3.4-13 2021-06-18 [2]
#> Rcpp 1.0.7 2021-07-07 [2]
#> readr 1.4.0 2020-10-05 [2]
#> readxl 1.3.1 2019-03-13 [2]
#> remotes 2.4.0 2021-06-02 [2]
#> rgdal 1.5-23 2021-02-03 [2]
#> rgeos 0.5-5 2020-09-07 [2]
#> rio 0.5.27 2021-06-21 [2]
#> rlang 0.4.11 2021-04-30 [2]
#> rmarkdown 2.9 2021-06-15 [2]
#> rnaturalearth 0.1.0 2017-03-21 [2]
#> rnaturalearthhires 0.2.0 2021-07-13 [2]
#> rpart 4.1-15 2019-04-12 [2]
#> rprojroot 2.0.2 2020-11-15 [2]
#> rstatix 0.7.0 2021-02-13 [2]
#> rstudioapi 0.13 2020-11-12 [2]
#> sass 0.4.0 2021-05-12 [2]
#> scales 1.1.1 2020-05-11 [2]
#> sessioninfo 1.1.1 2018-11-05 [2]
#> sf 1.0-0 2021-06-09 [2]
#> smatr 3.4-8 2018-03-18 [2]
#> sp 1.4-5 2021-01-10 [2]
#> sparr 2.2-15 2021-03-16 [2]
#> spatstat 2.2-0 2021-06-23 [2]
#> spatstat.core 2.2-0 2021-06-17 [2]
#> spatstat.data 2.1-0 2021-03-21 [2]
#> spatstat.geom 2.2-0 2021-06-15 [2]
#> spatstat.linnet 2.2-1 2021-06-22 [2]
#> spatstat.sparse 2.0-0 2021-03-16 [2]
#> spatstat.utils 2.2-0 2021-06-14 [2]
#> stringi 1.7.3 2021-07-16 [2]
#> stringr 1.4.0 2019-02-10 [2]
#> systemfonts 1.0.2 2021-05-11 [2]
#> tensor 1.5 2012-05-05 [2]
#> testthat 3.0.3 2021-06-16 [2]
#> textshaping 0.3.5 2021-06-09 [2]
#> tibble 3.1.3 2021-07-23 [2]
#> tidyr 1.1.3 2021-03-03 [2]
#> tidyselect 1.1.1 2021-04-30 [2]
#> tigris 1.4.1 2021-06-18 [2]
#> units 0.7-2 2021-06-08 [2]
#> usethis 2.0.1 2021-02-10 [2]
#> utf8 1.2.2 2021-07-24 [2]
#> uuid 0.1-4 2020-02-26 [2]
#> vctrs 0.3.8 2021-04-29 [2]
#> viridisLite 0.4.0 2021-04-13 [2]
#> withr 2.4.2 2021-04-18 [2]
#> xfun 0.24 2021-06-15 [2]
#> yaml 2.2.1 2020-02-01 [2]
#> zip 2.2.0 2021-05-31 [2]
#> source
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> local
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> Github (ropensci/rnaturalearthhires@2ed7a93)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#> CRAN (R 4.1.0)
#>
#> [1] /private/var/folders/mz/dw9fxm812gd3r6zjnv1rkmrr0000gn/T/Rtmpu0UTWu/temp_libpath1397a3804f8a8
#> [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/libraryThe current Git commit details are:
# what commit is this file at?
if ("git2r" %in% installed.packages() & git2r::in_repository(path = ".")) git2r::repository(here::here())
#> Local: main /Users/bocinsky/git/publications/p3k14c
#> Remote: main @ origin (https://github.com/people3k/p3k14c)
#> Head: [e39b168] 2021-07-13: updated readme as per reviews